CSV Files in Analytics: Taming the Variability
Written by Dan Homola |

Table of Contents
There are few formats as ubiquitous as CSV: most applications for which it makes even a smidge of sense to do so, support storing their output as CSV. Apart from its popularity, the format itself has quite a few additional advantages:
- It is human-readable, making it easy to create, read, and edit in any text editor, over the terminal, etc.
- It is approachable even for non-technical users, they can make sense of the raw contents easily.
- It can be versioned using Git and other version control systems.
- It is relatively condensed compared to other text-based formats like XML or JSON.
As with all things, CSV has its downsides, too:
- It is less efficient to store. For example, numbers take up space for each digit instead of being stored as a number.
- Being row-based, it is quite hard to get data only for certain columns: the whole file needs to be read even if we care about the first two columns, for example.
- There is no one universal CSV standard, it has several variants or dialects.
When adding support for CSV files into Longbow – our framework for creating modular data services – it was the last point that was especially challenging. In this article, we describe the approach we took with it.
What information do we need to extract
Let’s discuss what aspects of the CSV files we need to concern ourselves with when ingesting them into Longbow for further use. For each file, we need to derive the following:
- The encoding used by the file (ASCII, UTF-8, etc.).
- Dialect used by the file (delimiters, quotes, etc.).
- Names of the columns.
- Types of the data in the columns (integer, string, date, etc.).
- Preview of the first several rows so that the user can verify the CSV was parsed correctly.
We will explore the steps we took for each of these items in more detail in the rest of the article.
The ingest process
Before diving into the individual steps, let’s take a look at what the process of adding a new file looks like. First, the user uploads the CSV file they want to use to what we call a staging area. This is so we can run some analysis on the file using Longbow and show the results to the user. The user can review that the file is parsed correctly, and they can tweak some of the settings. Then, if they are satisfied with the results, they can proceed with confirming the file import. Once they do that, the file is moved from the staging area to the production area and it is then ready for use.
Storing the metadata
CSV has no dedicated way of storing any kind of metadata in the file itself (apart from somehow including it before the actual data), and we also want to support read-only input files. We had to devise a mechanism to store the metadata detected in the steps described below somewhere. We ended up with dedicated manifest files. The manifests are located right next to the relevant CSV files and have the same name with the .manifest suffix. They contain JSON-serialized versions of all the configurations we have collected both from the analysis and the user. Every time a particular CSV file is requested, we first check the manifest and use the configuration stored there to read the actual CSV file.
The configuration itself consists of options accepted by the Arrow CSV module (ReadOptions, ParseOptions, and ConvertOptions) that are used as-is when reading the CSV file. We also store information about date formats for any columns that should be interpreted as dates (more on that later).
Detecting the encoding
The very first step when reading an unknown CSV file (or any text file for that matter) for any analysis is to determine the encoding used by the file. This is to avoid any surprises with non-UTF-8 files being interpreted the wrong way. We use the charset_normalizer package for this purpose. The detected encoding is then used in subsequent reads of the CSV file.
Detecting the dialect and column names
The next step is to detect the so-called dialect of the CSV file. The dialect describes some of the structural properties of the CSV:
- What is the separating character for the individual columns?
- Are there any quotation marks used to escape the separators, and if so, how can they be escaped?
We also need to detect the column names. Some CSV files store the column names in the first row, some do not store them at all, and we need to generate some ourselves.
We use DuckDB's sniff_csv function to gather all of this information. It gives us all the structural information about the file, like the delimiters, quotes, etc. It also detects the column headers if there are any, falling back on autogenerated column names. You can read more about the DuckDB CSV capabilities in their introductory blog post. We also need to make sure that the file we feed into DuckDB is in UTF-8. Otherwise, it fails. We make use of the detected encoding and prepare a special copy of the input file just for DuckDB in case the original is not in UTF-8 (or ASCII).
def _detect_dialect_and_header_and_column_names(
sample_filename: str,
encoding: str,
) -> tuple[CsvDialect, int, list[str]]:
needs_conversion = encoding not in ["utf_8", "ascii"]
if needs_conversion:
duckdb_input_file = sample_filename + ".utf_8.csv"
# ... convert the sample file to utf-8
else:
duckdb_input_file = sample_filename
try:
return _run_duckdb_detection(duckdb_input_file)
finally:
if needs_conversion:
os.unlink(duckdb_input_file)
def _run_duckdb_detection(
duckdb_input_file: str,
) -> tuple[CsvDialect, int, list[str]]:
# use only one thread, we will always run only one query at a time from this
conn = duckdb.connect(":memory:", config={"threads": 1})
query = conn.execute(
"SELECT Delimiter, Quote, Escape, HasHeader, Columns FROM sniff_csv(?)",
[duckdb_input_file],
)
query_result = query.fetchone()
if not query_result:
raise ValueError("Unable to detect file dialect.")
(delimiter, quote, escape, has_header, encoded_columns) = query_result
# the detection may return \x00 as a delimiter, need to normalize to None
dialect = CsvDialect(
delimiter=delimiter if delimiter != "\x00" else None,
quotechar=quote if quote != "\x00" else None,
escapechar=escape if escape != "\x00" else None,
)
# duckdb returns the columns as string, we have to directly inject it back
# into duckdb to parse it correctly
col_query = conn.execute("SELECT a.* FROM (SELECT " + encoded_columns + " as a)")
decoded_columns = col_query.fetch_arrow_table().column_names
if has_header:
# the header takes as many rows as the column with the most newlines in the name
header_row_count = max(col_name.count("\n") for col_name in decoded_columns) + 1
else:
header_row_count = 0
return dialect, header_row_count, sample_filenameBefore the sniff_csv was available, we used the CleverCSV library for this step. Still, the DuckDB variant performs better (we observed a ten-fold improvement in the overall time) and allowed us to simplify the code since it can detect the dialect and column names in one step.
Detecting the data types
Having a way to read the file with the schema in hand, we can proceed with determining the actual data type of each column. You might ask, “Why not use the types detected by DuckDB?” or “Why not use the automatic detection that Arrow CSV has?”. There are a few reasons, but the most significant one has to do with the various date formats we want to support.
The DuckDB CSV sniffer only supports one date format per file, so if you use one date format in one column and another format in another column, it will not work. Arrow CSV does support different date formats per column, but the set of date formats it supports is limited. While it would work great with ISO 8601 compliant dates, for example, it would not recognize strings like:
- Jan 22, 2023 01:02:03
- 01 22 23 01:02:03
- 20230122
as potentially being dates as well. This is not to say the Arrow detection is wrong (after all, the last example may very well be just an integer). We just need to support a wider set of formats.
You can specify which date formats you want Arrow to try, but in case of ambiguity, it will always assume that the first matching format is correct. We want our users to disambiguate the date format manually: only they know which format is the correct one.
Another limitation of the Arrow CSV approach is that you either get the most precise data type detection (but you need to read the whole file into memory -which obviously does not scale that well), or you can use the batch-based approach. Still, only the first batch of the file is used for the data type detection making it less precise.
We want the most precise detection while conserving the memory. To that end, our pipeline is constructed a bit differently. First, we tell Arrow to read the file batch by batch and to treat each column as a string so that we avoid any automatic detection performed by Arrow. This is where the column names come in handy: you need their names to reference them in the Arrow CSV options. Next, we pipe this source into a custom Acero pipeline that allows us to run the analysis extremely quickly on the entire file in a streaming fashion, keeping the memory footprint small.
Acero streaming engine
What is Acero, you might wonder. Acero is an experimental streaming engine for running queries on large data. In Acero, you specify the processing pipeline declaratively, using several building blocks like projections, filters, and aggregations. You can choose from a wide range of predefined compute functions and crucially, you can also define your own custom functions (User Defined Functions, UDFs for short). The UDFs are fairly easy to write: you worry only about the transformations you want to perform. Acero figures out the rest. What’s more, you can use several languages to do so, we use Python for the data type detection pipeline and Cython for the pipeline we use to read the CSV data using the detected types. If SQL is more up your alley, you can use Substrait to generate the Acero query plan from an SQL query.
The type detection pipeline
From a high-level perspective, our type detection pipeline is very simple: it has one source node reading the CSV file and one projection node running the UDF detection algorithm. Ideally, there would also be an aggregation node at the end that would aggregate the results of each projection batch. Unfortunately, Acero does not seem to support UDFs in the aggregation nodes yet, so we run the aggregation in pure Python.
The detection UDF is run in parallel for every column in isolation and works like this. For each batch of values in a column:
- We detect which values are null or empty - we use regular expressions
import pyarrow.compute as pc
is_boolean_vec = pc.match_substring_regex(
array,
# values taken from the defaults in pyarrow.csv.convert_options
pattern=r"^$|^(true|false|0|1)$",
ignore_case=True,
memory_pool=ctx.memory_pool,
)We use regular expressions and the strptime function to detect possible date formats (based on a set of supported date formats).
We return the following values
- All the types the values in the batch conform to order by the specificity (e.g. integer is more specific than a double).
- All the date formats that can be used to parse all non-empty values in the batch as a valid date.
- Whether any of the values in the batch is null or empty.
- Whether all of the values in the batch are null or empty.
We then aggregate the results for all the batches for each column so that we get the final result:
- The most specific type usable for the column.
- All the date formats that can be used to parse all the non-empty values in all the batches.
- A flag indicating whether the column is nullable: i.e., it contains at least one value that is null or empty.
Reading a preview
To allow the user to make an informed decision whether we “understood” the file properly and to allow them to pick the correct date format from those that we detected as suitable, we read a small sample of the data using the options we intend to use once the file is confirmed by the user. We return this preview as a part of the response, along with all the options and configurations we detected.
You might wonder, “Why does the user need to pick a date format?”. This is to handle situations where the date values are ambiguous. Imagine a file that only has these two values in a column: 01/01/2024 and 01/02/2024. Do these correspond to January 1st and 2nd? Or are they January 1st and February 1st? Only the user knows which is the case, so in these (admittedly rare) cases, they need to pick the correct date format for us to use.
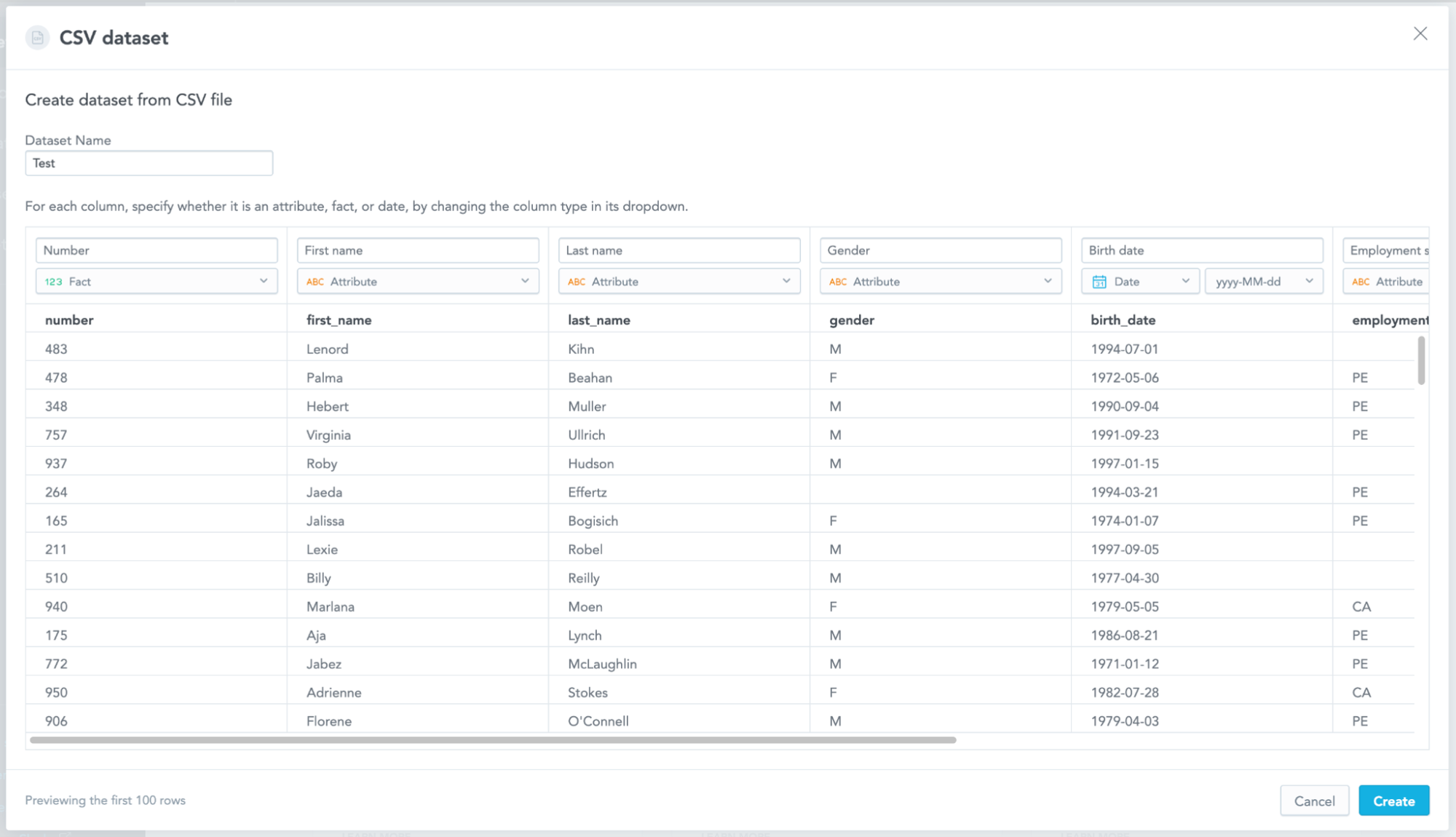
Using the CSV file as a source of data
Once the user confirms the CSV file is correctly parsed, the file is moved to the production area of the file storage, and a manifest file with all the metadata is created. When there is a computation run that needs to access the CSV data, it uses the metadata in the manifest to prepare a RecordBatchReader that uses another Acero pipeline with another UDF for reading the date columns using the correct date format. The UDF is a thin wrapper around the strftime function written in Cython that does not fail on empty values but fails on invalid non-empty values. The default strftime either fails on empty values or returns null for anything it cannot parse, neither of which is what we want.
The resulting RecordBatchReader can then be consumed by the rest of Longbow, business as usual. There is a dedicated article coming about that particular part of Longbow, so stay tuned!
Summary
CSV files are one of the most used formats for storing structured data. Their relative looseness and simplicity make them easy to produce, but they are also quite challenging to read and parse automatically. We have outlined the way we do it for Longbow, leveraging the DuckDB CSV sniffing functionality and the Apache Arrow capabilities: its CSV module and the Acero streaming engine.
Want to learn more?
As always, I’m eager to hear what you think about the direction we are taking! Feel free to reach out to us on the GoodData community Slack.
Want to try it out for yourself? Consider using our free trial. Want to learn what else we are cooking at GoodData? Join GoodData Labs!
Written by Dan Homola |

